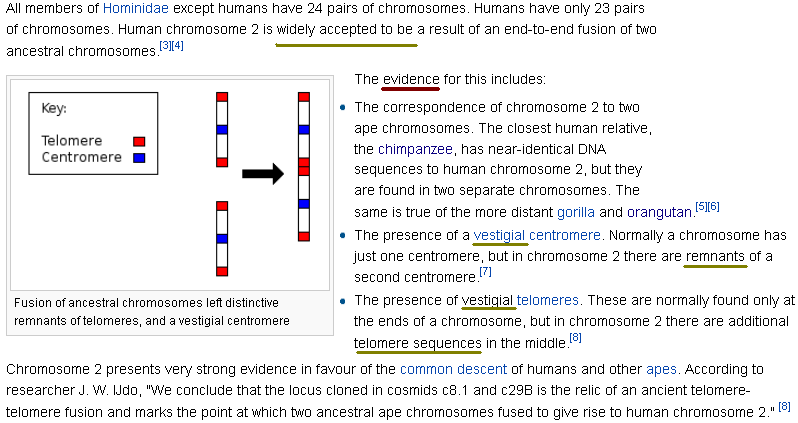
To solve the problem of different number of chromosomes between apes and humans, Darwinists suggested the ‘chromosome 2 fusion model’. This scenario involves the claim that the fusion of two small chimpanzee-like chromosomes (2A and 2B) formed one chromosome in humans, leading to the difference in diploid chromosome numbers between humans and great apes. While the chromosome 2 fusion model is routinely touted as dogma, very little new genomic data, although readily available for analysis, has been presented as evidence.
Wikipedia mentioned that the model is "
widely accepted":
But, What you need to know is:
1- A majority of the data for the fusion model is based on DNA
hybridization and chromosomal
staining experiments conducted prior to the sequencing of the human and chimpanzee genomes.
2-
Synteny (shared ordering of genes between organisms) of the chromosomes is not a surprise. We already knew there was a vast amount of similarity between humans and primates both in terms of physical characteristics and genetic material and structure. It is a mistake to assume that observing similarities necessarily brings you to the conclusion of common descent. Taxonomy based on physical characteristics was already a very well established science when the idea of common descent came on the scene.
3- The telomere region involves a complex and dynamic framework of DNA motif repeats 5’ to 3’ (TTAGGG)n , structural loops, structural and a wide variety of proteins. and it confer stability and preventing fusion or shortening of the chromosomes. It's in perfect tandem of DNA from about
10 to 15 kb (10,000 to 15,000 bases) and contains
1,667 to 2,500 telomere repeats at each chromosome end.
4- In a head-to-head fusion of two chromosomes,
we would expect at least 5,000 bases of (TTAGGG)n repeats in tandem, we would also expect the orientation of the plus-strand repeat to change to the reverse complement (CCCTAA)n, which should also occur in near-perfect tandem for approximately 5,000 or more bases. The area containing the suspected ‘fusion region’ is often called
2qfus or 2chr2fus and occupies the genomic area
between 2q13 and 2q14.1.
5- Within the 10 to 30 kb window of DNA sequence surrounding the hypothetical fusion site,a glaring paucity of telomeric repeats exist that appear mostly as
independent monomers, not tandem repeats. The TTAGGG repeat to the left of the fusion site, less than
35 motifs exist. For the CCCTAA reverse complement sequence, to the right of the fusion site, less than
150 telomere motifs can be found.
6- The only research group to seriously analyze the actual fusion site DNA sequence data in detail were confounded by the results which showed a lack of evidence for fusion—a genomic condition for this region which they termed ‘
degenerate’.(Fan, Y. et al., Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes, Genome Research 12:1651–1662, 2002. )
7- Fairbanks said that
44 out of 158 repeats match (28%) and that the rest of the sequences are ‘close’. The problem is, to obtain even this low match level, the consensus reading frame is entirely ignored and ambiguous matches are contrived by assuming many insertion and deletion mutations of varying sizes.
8- There are
genes throughout the alleged fusion region. In an analysis of a
614 kb area encompassing the postulated chromosome fusion site, Fan et al. found evidence of “at least
24 potentially functional genes and
16 pseudogenes”.(Fan, Y. et al., Gene content and function of the ancestral chromosome fusion site in human chromosome 2q13-2q14.1 and paralogous regions, Genome Research 12:1663–1672, 2002.). In the
30-kb region directly encompassing the fusion site, which should definitely be devoid of any genes, there exists
two actively transcribed genes, each in a flanking position in regard to the fusion site (one on each side). There are also at least
two other genes in the immediate vicinity of the fusion site thought to be inactive due to frame shift mutations. However, research related to the human ENCODE (Encyclopedia of DNA Elements) project has shown that many genes thought to be inactive (pseudogenes) are actually functional due to a variety of newly discovered regulatory mechanisms.(Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project, Nature 447:799–816, 2007)
9- If the telomere motifs that populate internal areas of chromosomes serve some important, yet unknown function, the chromosome fusion model actually impedes research aimed at determining possible function in these regions.
10- telomeres are designed to
prevent fusion. Broken chromosomes at any location immediately invoke the cell’s double-stranded DNA repair machinery where the aberrant fusion of fragments actually triggers cell fault tolerance mechanisms.6 In the case of an aberrant fusion, a senescence response or programmed cell death (
apoptosis) cascade is normally triggered, effectively eliminating the damaged cell from the system.
A cell with telomeres that have progressively shortened over time and reached a threshold length will also activate the double-stranded DNA repair machinery; inducing cell senescence and/or death. When in certain types of germline cells, telomerase adds telomere repeats to shortened telomeres, chromosomes are ‘healed’ and can again become stable. The telomeres cap the ends of linear chromosomes and effectively prevent fusion or trigger cell elimination if the telomere is shortened to a certain point, damaged, or aberrantly fused.6 According to the fusion model, this protective process was somehow bypassed in early humans.
11-
Telomere sequence is not unique to the telomere. the existence of telomeric repeats in internal sites is not unusual or unexpected. We know that ITS (interstitial telomeric sequences) present in Chromosomes 1,4,5,9,12,16, and 17 (
http://www.ncbi.nlm....4?dopt=Abstract)
12- The evidence for a second remnant centromere at any stage of sequence degeneracy is negligible.The supposed evidence includes the finding that “every human and great-ape chromosome centromere contains a highly variable DNA sequence that is repeated over and over, a 171 base-pair sequence called the Alphoid sequence.”3 Fairbanks adds that scientists have “searched for Alphoid sequences in human chromosomes and found them at every centromere, as expected. They also found Alphoid sequences at the site in human chromosome 2 where the remnants of this second centromere should be. These remnants are evidence of a now-defunct centromere.” problem is that, although research has been done on some primates, no systematic study of centromeres exists to determine how common alphoid DNA is in mammals.(Baldini, A. et al., An alphoid DNA sequence conserved in all human and great ape chromosomes: evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13, Human Genetics 90:577–583, 1993) Baldini et al. found that the “highest sequence similarity between human and great ape alphoid sequences is 91%, much lower than the expected similarity for selectively neutral sequences.”
13-
Alpha-satellite DNA or alphoid DNA, although found in centromeric areas, is not unique to centromeres and is also highly variable. Because highly variable alphoid DNA is also commonly found in non-centromeric regions of human chromosomes, their presence does not indicate the remnants of a degenerate centromere. Based on the reasoning of Fairbanks and others promoting the human chromosome 2 fusion model, one could conclude that human chromosomes contain literally hundreds of degenerate centromeres.
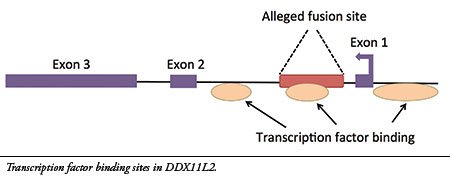
14- The site is located inside a gene called
DDX11L2 on human chromosome 2. Furthermore, the alleged fusion sequence contains a functional genetic feature called a “transcription factor binding site” that is located in the first intron (non-coding region) of the gene (see illustration). Transcription factors are proteins that bind to regulatory sites in and around genes to control their function, acting like switches. The
DDX11L2 gene has three of these areas, one of which is encoded in the alleged fusion site.(
source )